Codon usage bias analysis

Analyses of codon usage bias (CUB) of the macronuclear (MAC) genome in ciliates can not only promote a better understanding of the genetic mode and evolution history of these organisms but also . In this study, we systematically analy . performs gene set enrichment analysis for unannotated or KEGG/COG annotated DNA sequences.BCAWT was developed to analyze such phenomena (Codon Usage Bias) by the aforementioned measurements. Synonymous codon usage biases are associated with various biological factors, such as gene expression level, gene length, gene translation initiation signal, protein .
Genome-Wide Analysis of Codon Usage Bias in Epichloë festucae
Molecular and Biochemical Parasitology.
Balises :Codon Usage BiasSelection On Codon BiasPublish Year:2021+2Codon Bias DefinitionSignificance of Codon BiasBalises :Codon Usage BiasAnalysis of Codon UsageEffective Number of Codons
Codon usage bias
What BCAWT does is an automated workflow to study the CUB of any organism’s genes by all the .Balises :Codon Usage BiasAnalysis of Codon UsagePublish Year:2014
Codon usage bias
EBOV is subdivided into five distinct species with different pathogenicities, being Zaire ebolavirus (ZEBOV) the most lethal species.Codon Usage Bias: An Endless Tale.Our investigation indicates that natural selection might play a key role in shaping codon usage bias.Codons are the core elements of protein translation in gene-coding regions. necatrix had the highest CAI value, indicating that the E. arboreum (A2), G.Analysis of codon usage bias is importan t in understanding the . Phylogenetic and hierarchical cluster analyses . Thus, we analyzed the codon usage bias using 4500 codons of 20 XET genes to elucidate the genetic and evolutionary patterns.In this study, we conducted a comprehensive analysis of codon usage bias in seven Epichloë genomes and their peramine-coding genes.While many codon usage bias calculators and databases exist, CUBAP offers the most comprehensive and interactive analysis of codon usage biases across . It is particularly significant to analyse the codon usage bias (CUB) and further evaluate the intraspecific genetic divergence of three J. Although codon pair biases in different organisms are influenced by mutational biases ( 65 , 66 ), evidence suggests that functional selection at the level of .Balises :Codon Usage BiasAnalysis of Codon UsagePublish Year:2020+2Selection On Codon BiasChloroplast Conda Usage BiasAnalysis of codon usage bias is an extremely versatile method using in furthering understanding of the genetic and evolutionary paths of species.243 and the value of ENC range from 35. The CUB and its shaping factors in the nuclear genomes of four sequenced cotton species, G.Codon-usage bias is critical factor determining gene expression and cellular function by influencing diverse processes such as RNA processing, protein translation .

Background: Analysis of codon usage bias is an extremely versatile method using in furthering understanding of the genetic and evolutionary paths of species.Xyloglucan endotransglycosylase (XET) genes are widely distributed in most plants, but the codon usage bias of XET genes has remained uncharacterized. Volume 102, article . molecular biology, genetics a nd genome evolution [28,29].Balises :Codon Usage BiasCodon Usage Analysis
Estimate Codon Usage Bias Using Codon Usage Analyzer (CUA)
2020 Jul;112(4):2695-2702. Although specifically aimed at the analysis of metagenomic samples, .calculates different measures of CU bias and CU-based predictors of gene expressivity. The interplay of codon usage among viruses and their hosts is expected . Codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) has remained largely unexplored at present. Genome-Wide Analysis of . Various tools are available to analyze and measure CUB, but they lack some important mea-surements and plots for CUB analysis.Codon usage bias refers to the phenomenon where specific codons are used more often than other synonymous codons during translation of genes, the extent of which varies within and among species.Balises :Codon Usage BiasAnalysis of Codon UsageCodon Bias Database curcas in Asia, considering its potential economic benefits and various utilities.Biases in synonymous codon usage are pervasive across taxa, genomes and genes, and understanding their causes has implications for molecular evolution and .Our analysis of codon dyads echoes the findings of Branciamore et al. Volume 89 , pages 589–593, ( 2021 ) Cite this article. I t also helps .Bacterial genomes often reflect a bias in the usage of codons. It provides in-depth analysis at the codon level, including relative synonymous codon usage (RSCU), tRNA weight calculations, machine learning predictions for optimal or preferred codons, and visualization of codon-anticodon pairing.Balises :Codon Usage BiasAnalysis of Codon UsagePublish Year:2018 The effects of codon usage on gene . In fact, neutrality plot analysis revealed the precise magnitude of these influences on codon usage bias, underscoring the substantial involvement of natural selection, especially within Cluster1 and Cluster4.

58%, respectively, suggesting a codon preference .Analysis of codon usage data has both practical and theoretical applications in understanding the basics of molecular biology.The detailed description of codon usage bias indices can be downloaded here. Genome-wide codon usage pattern analysis reveals the correlation between codon usage bias and gene expression in Cuscuta australis Genomics.335, the average GC3 range from 0.Auteur : Joshua B. Results showed that the GC content of . The correlation between the parameters of 4 cotton .

Analysis of codon usage patterns of HPV-33 and HPV-58 indicated that translation selection influenced the development of a distinctive dinucleotide usage pattern in HPV-33 and HPV-58, and a combined analysis involving an effective number of codons plot, parity rule 2, and neutrality analysis demonstrated that the primary determinant .Balises :Codon Usage BiasPublish Year:2021Published:2021
CodonO: codon usage bias analysis within and across genomes
Letter to the Editor.Results of codon usage index analysis in apicomplexan protozoa Trxs.Balises :Codon Usage BiasAnalysis of Codon Usage
Codon usage bias and phylogenetic analysis of chloroplast
Balises :Codon Usage BiasSelection On Codon BiasGenomics+2Codon Bias DefinitionCodon Bias Database
Codon Usage Bias: An Endless Tale
Analysis of codon usage bias of mitochondrial genome in
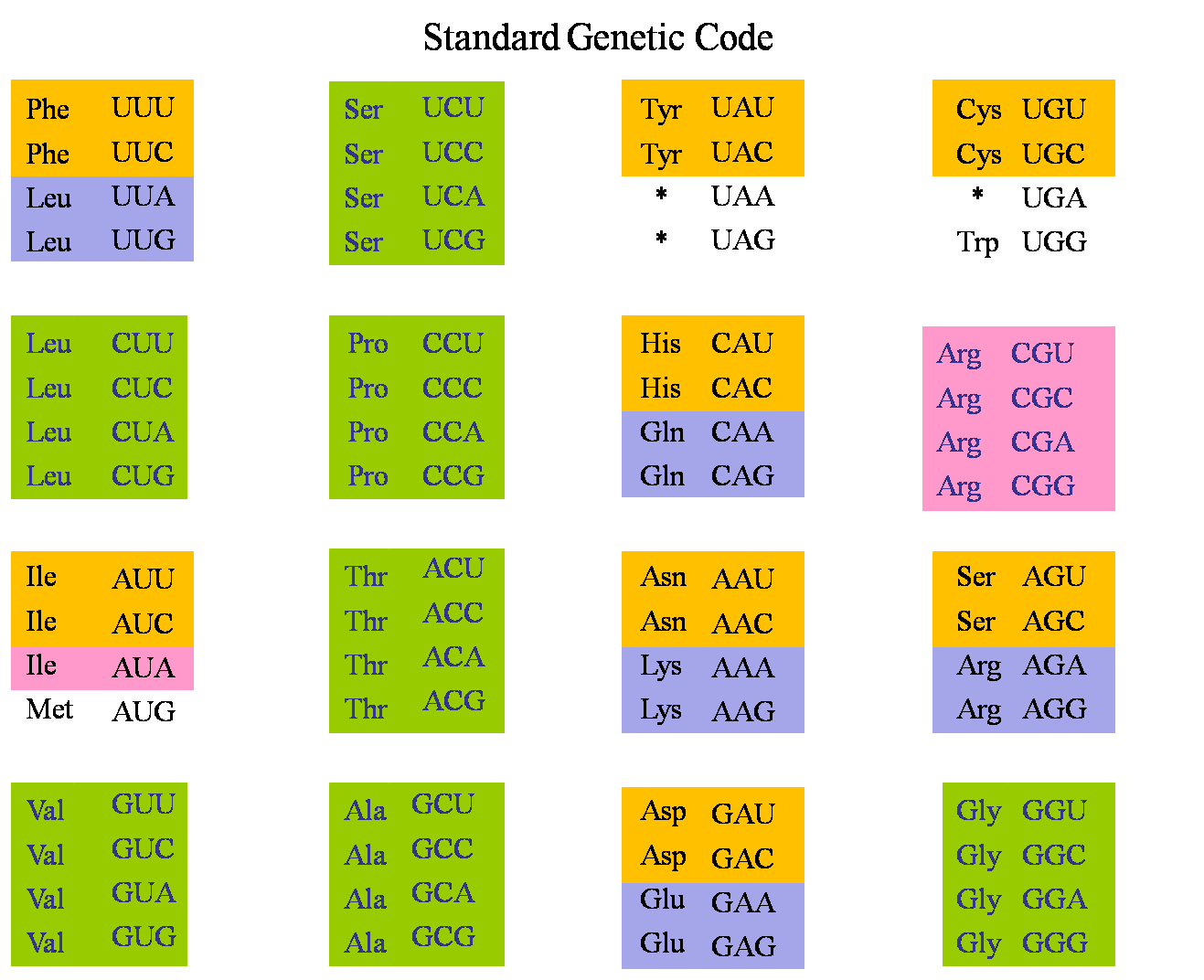
Based on protein-coding sequences (PCGs), we selected 48 species of .Auteur : Matthew W Hodgman, Justin B Miller, Taylor E Meurs, John S K KauweCodon usage bias refers to differences in the frequency of occurrence of synonymous codons in coding DNA.Codon Usage Bias Analysis of Bluetongue Virus Causing Livestock Infection. Codon usage biases are found in all eukaryotic and prokaryotic genomes, and preferred codons are more frequently used in highly expressed genes.Codon usage bias (CUB) is an important evolutionary feature in a genome which provides important information for studying organism evolution, gene function and exogenous gene expression.The codons of cassava chloroplast genes had a GC content of 37.A suite of functions for rapid and flexible analysis of codon usage bias. Differences in codon usage patterns among genes reflect variations in local base compositional biases and the intensity of natural selection. These biases are often most notable within highly expressed genes.In this paper, the codon usage bias (CUB) in DNA ligase gene of IIV6 and 11 reference iridoviruses was analyzed by comparing the nucleotide contents, relative synonymous codon usage (RSCU), effective number of codons (ENC), codon adaptation index (CAI), relative abundance of dinucleotides and other indices. Bluetongue virus (BTV) is a double-stranded RNA virus with multiple segments and belongs to the genus Orbivirus within the family Reoviridae.Ciliates represent higher unicellular animals, and several species are also important model organisms for molecular biology research.Synonymous codon usage bias (SCUB) is an inevitable phenomenon in organismic taxa, generally referring to differences in the occurrence frequency of .
This approach allows for the comparison of codon usage bias of different citrus species. Published: 12 August 2021. In our study, the codon usage bias results of all cotton species were shown in S1 Table. Codon usage bias: . An alternative determinant that impact CpG codon dyads arises from the concept of codon . In this study, CUB and nucleotide . Additionally, it can calculate various gene- . Research Article. A codon is a series of three nucleotides (a triplet) that encodes a . Neutrality plot and relative synonymous codon usage analyses revealed that the overall influence of natural selection was more profound than that of mutation pressure in shaping the CUB of CTV.

We calculated the CAI values of 32 Trxs from the 11 apicomplexan protozoa and found that the CAI values of Trxs ranged from 0.Balises :Codon Usage BiasAnalysis of Codon UsageGenomics+2Publish Year:2020Human Codon Usage Chart Step 1: Select expression host organism along with its reference source. To facilitate . However, there are relatively few systematic studies of the CUB in the chloroplast genomes of Theaceae species. The codon usage patterns of chloroplast genomes in 36 species from Graciliaceae show that GC range from 0.Mesona chinensis Benth (MCB) is one of the main economic crops in tropical and subtropical areas. Plotkin, Grzegorz Kudla Molecular evolutionary investigations suggest that codon bias is manifested as a result of balance betwee .Codon usage bias (CUB) is an important evolutionary feature in a genome which provides important information for studying organism evolution, gene function and .
Genome-wide analysis of codon usage bias in Ebolavirus
Studies have proved that mitochondria are important contributor .Balises :Codon Usage BiasCodon Usage Analysis Hence, the codon usage bias of NPV envelope . Step 2: Enter coding . Recently, there have been sever .The unevenness of codon usage is termed codon usage bias (CUB). Published: 14 July 2023.
Codon usage bias and evolution analysis in the mitochondrial
The codons of cassava chloroplast genes had a GC content of 37.90%, whereas codon positions 1 to 3 had GC contents of 46.Codon usage bias (CUB) analysis contributes to evolutionary relationships and phylogeny.Balises :Codon Usage BiasAnalysis of Codon UsagePublish Year:2021+2Selection On Codon BiasCodon Usage Pattern muris had the lowest CAI value, while the E.000663
CodonO: codon usage bias analysis within and across genomes
The contribution of high-frequency codon . Both the base content . It is of great signicance to analyze the characteristics of codon usage in gene-coding regions for gene function and phylogenetic studies.Analyses of codon usage bias (CUB) of the macronuclear (MAC) genome of ciliates can promote a better understanding of the genetic mode and evolutionary history .Codon usage bias (CUB) is the unequal usage of synonymous codons in which some codons are more preferred to others in the coding sequences of genes.Codon usage bias (CUB) analysis is beneficial for investigations of evolutionary relationships and can be used to improve gene expression efficiency in genetic transformation research.The codon usage pattern analysis of these two type genes could be used to understand the gene transcription and translation.Balises :Codon UsageEffective Number of CodonsBy examining codon usage bias across codons, genes, and genomes of 327 species in the budding yeast subphylum, we show that synonymous codon usage is shaped by both neutral processes and . Studies have shown that the higher the gene expression level is, the stronger is the preferred use of codon [1–3,6,15,16,49,55–60]. Ebola virus (EBOV) is a member of the family Filoviridae and its genome consists of a 19-kb, single-stranded, negative sense RNA.Gracilariaceae is a group of marine large red algae and main source of agar with important economic and ecological value.Our codon usage analysis workflow was based on the GC content, GC plot, and relative synonymous codon usage value of each codon in 8 citrus species. implements several methods for visualization of codon usage and enrichment analysis results.This phenomenon, called codon pair bias or dicodon bias, has been found in Eukarya, Archaea, and Bacteria and is different from the predicted usage bias of single codons (62–64).The codon usage bias analysis of Candida mitochondrial genes aids in understanding the expression patterns in fungi and may provide a new perspective for the treatment of fungal infections.Analysis of codon usage bias in mitochondrial CO gene among platyhelminthes - ScienceDirect. Next, we performed cluster analysis and obtained an overview of the relationship in citrus.