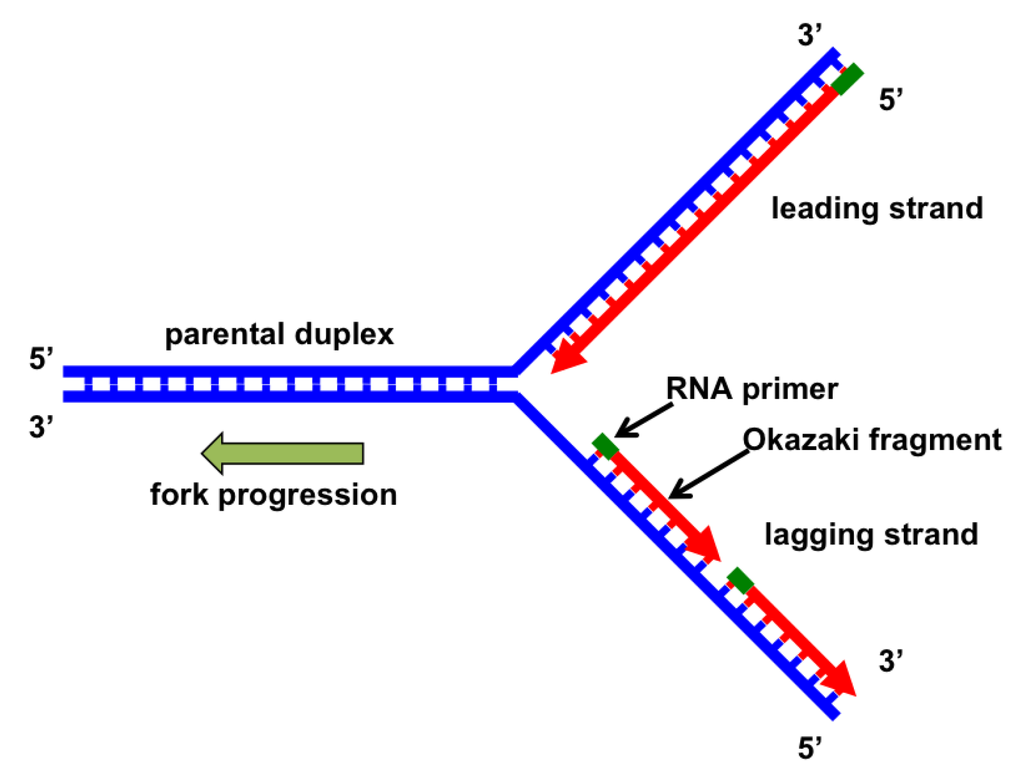
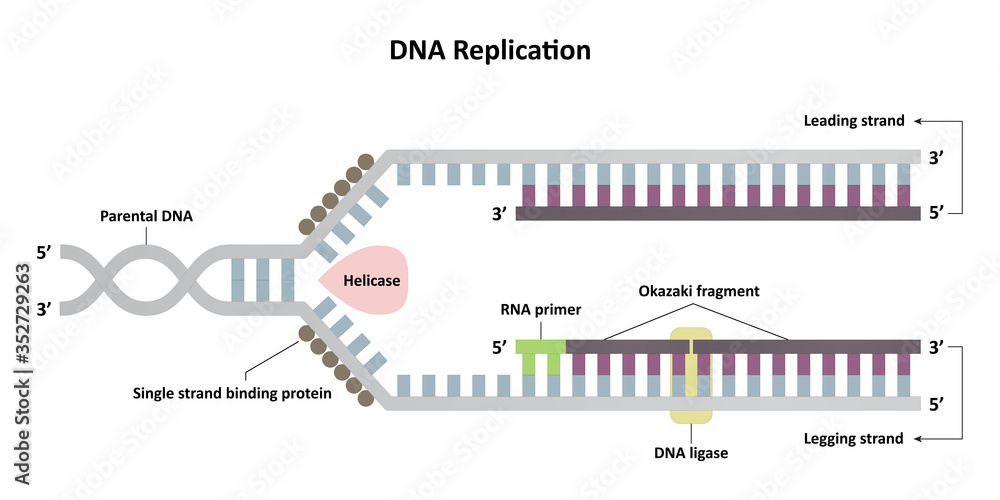
Dna polymerase iii lagging strand
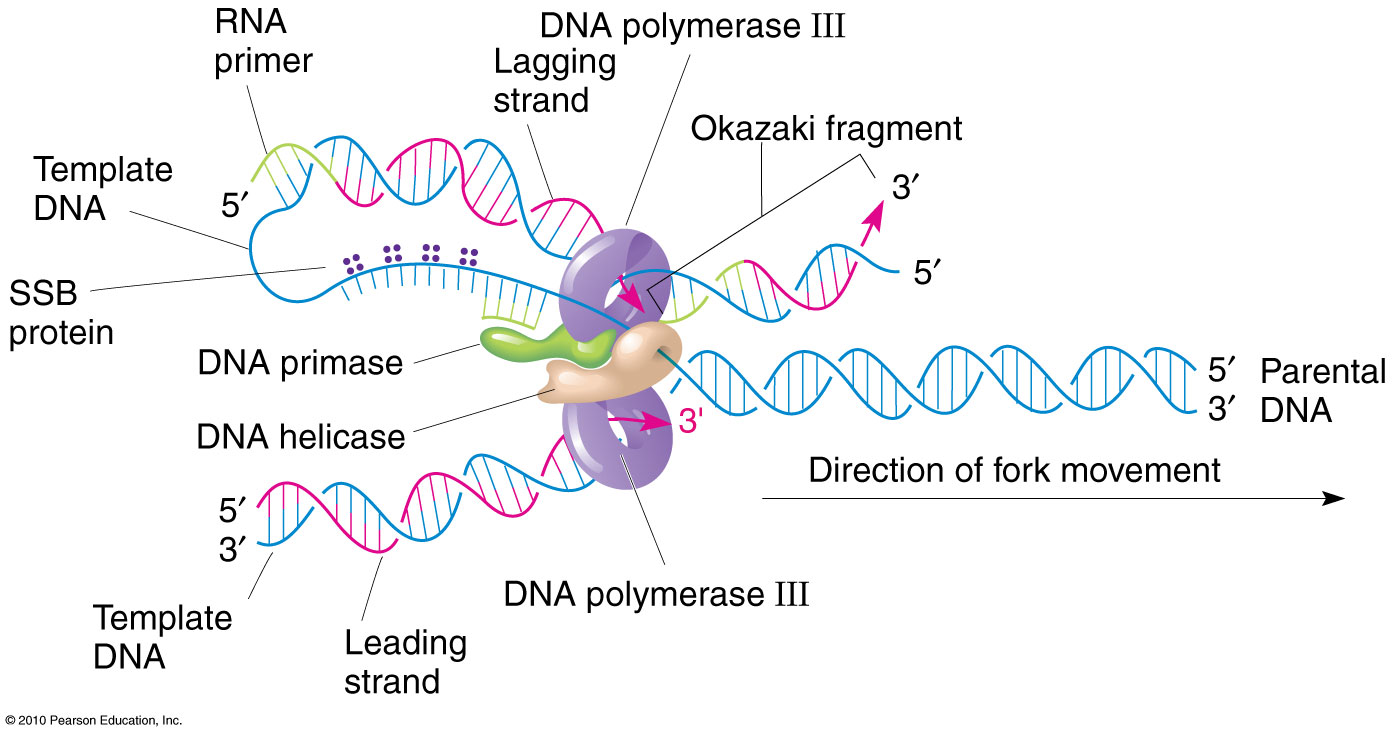
At least five prokaryotic DNA polymerases have been discovered to date with DNA Polymerase III (Pol III) being the primary polymerase for replication. coli, including DNA polymerase III (discovered in the 1970s) and DNA polymerases IV and V (discovered in 1999). Is a diagram of DNA replic. A protein called the sliding clamp .
Genome Replication
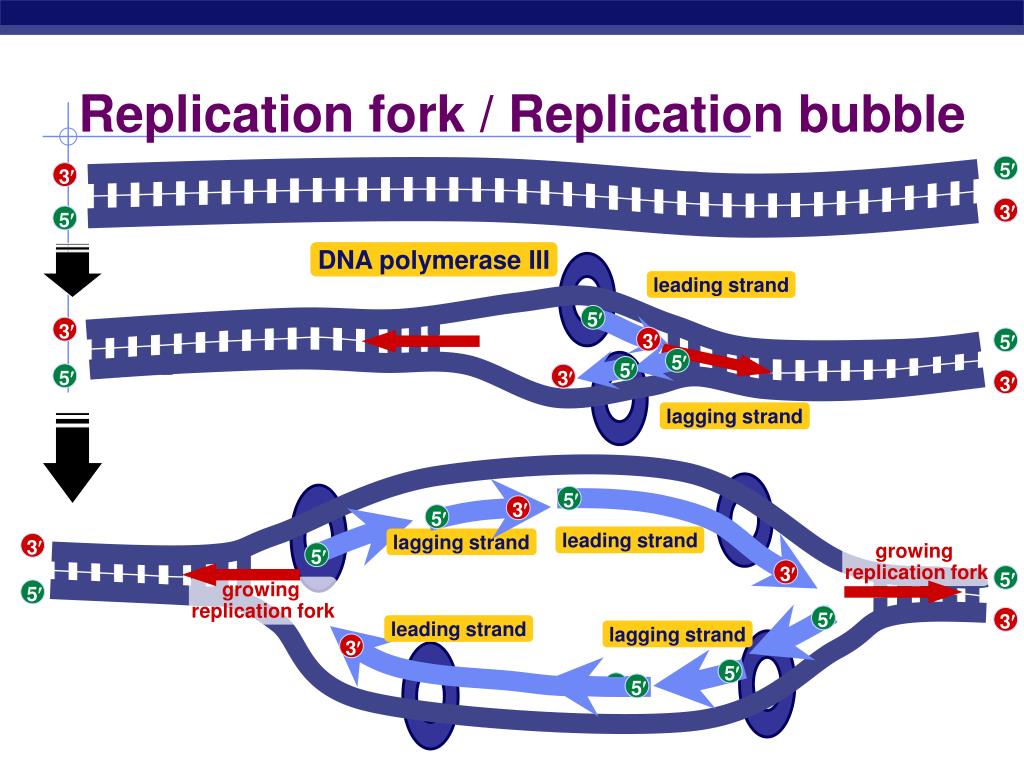
As the two parental (template) DNA strands separate at a replication fork, each of the strands is separately copied by a DNA polymerase III (orange), producing two new daughter strands (light blue), each complementary to its respective parental strand. This enzyme can .
Role of the Core DNA Polymerase III Subunits at the Replication Fork
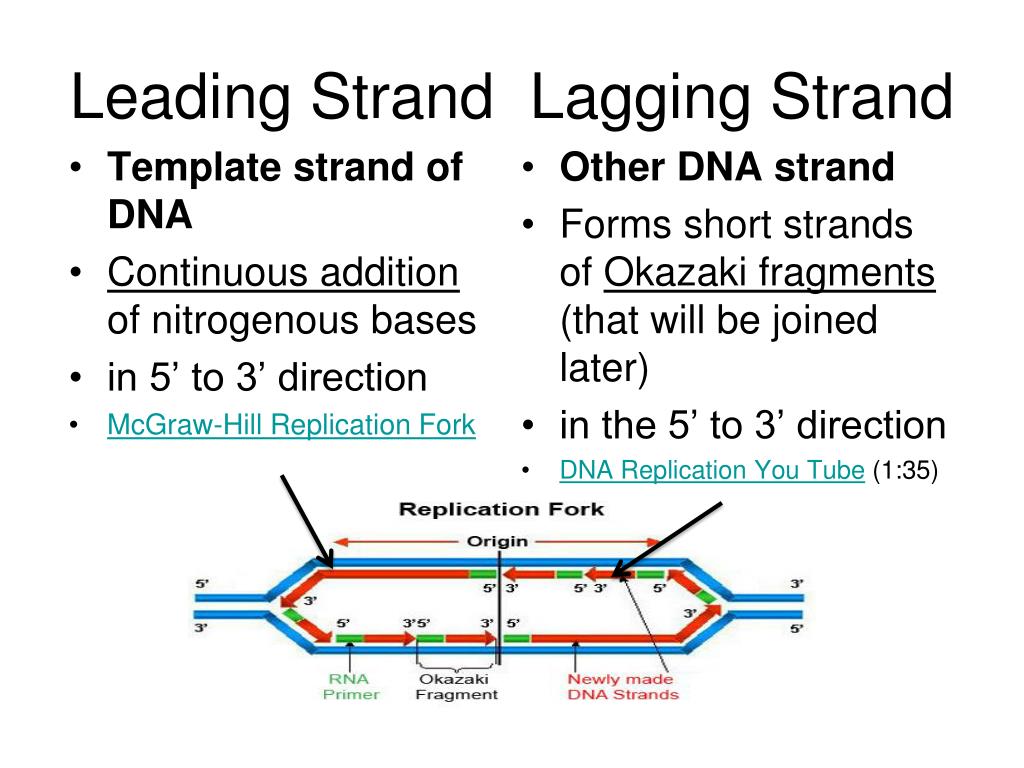
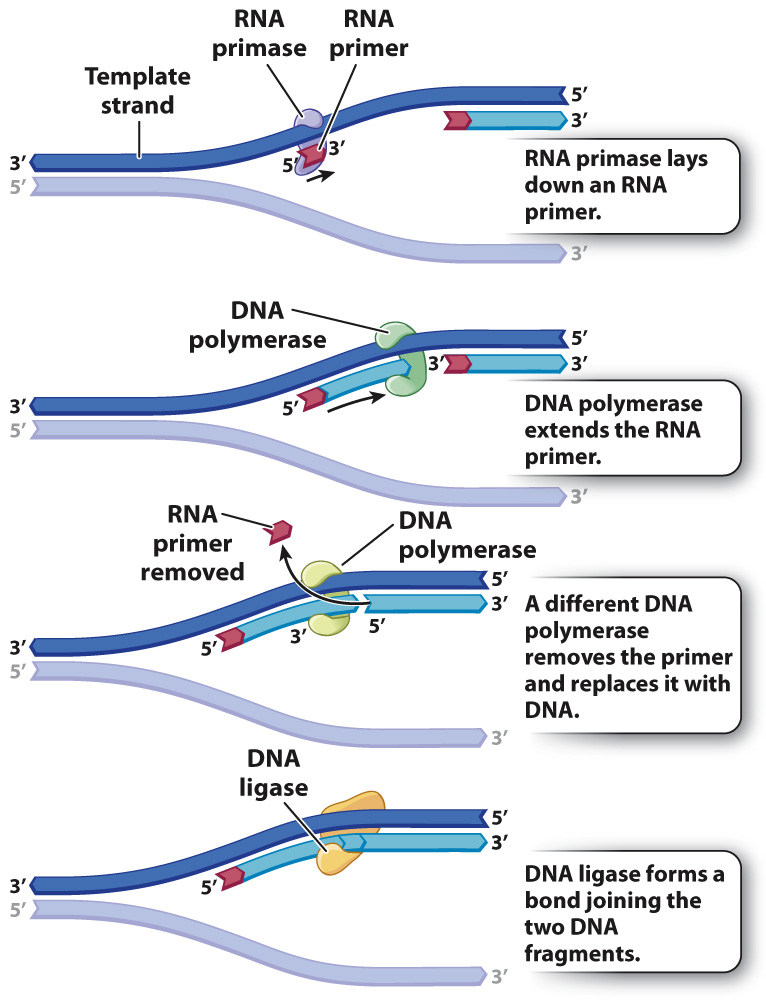
The primary enzyme involved in this is DNA polymerase which joins nucleotides to synthesize the new complementary strand. Pol I is used primarily to fill in gaps created during lagging strand synthesis (described below) or through error-correcting mechanisms. The DNA polymerase III holoenzyme is composed of 10 subunits. However, because the parental DNA strands are antiparallel, the lagging strand must be made in short fragments. Replication fork. DNA polymerase adds nucleotides to the deoxyribose (3’) ended strand in a 5’ to 3’ direction.Escherichia coli DNA polymerase III holoenzyme contains 10 different subunits which assort into three functional components: a core catalytic unit containing . An RNA primer complementary to the parental strand is synthesized by RNA primase and is elongated by DNA polymerase III through . Dynamics at the .As the chases increased in length, giving DNA more time to replicate, the lagging strand fragments started integrating into longer, heavier, more rapidly sedimenting DNA strands. DNA polymerase III uses this primer to synthesize the daughter DNA strand.DNA polymerase I aids in lagging strand synthesis as it removes the RNA primers and incorporates DNA in its place.DNA polymerase III is the main enzyme that then elongates new DNA. C) DNA polymerase II. Because the two parental strands are antiparallel, the two new strands (the leading and .Saccharomyces cerevisiae DNA polymerase δ (Pol δ) and DNA polymerase ε (Pol ε) are replicative DNA polymerases at the replication fork.As you have learned, the DNA polymerase enzyme can add nucleotides in only one direction.The leading strand is made continuously in a 5' to 3' direction by DNA polymerase III as the DNA helicase unwinds the parental DNA helix.
Leading vs Lagging DNA Strand: Difference and Comparison
The 3′
During DNA replication (copying), most DNA polymerases can “check their work” with each base that they add.The strand with the Okazaki fragments is known as the lagging strand.Each DnaB helicase binds to at least two molecules of DNA polymerase III (Pol III), which synthesize the leading and the lagging strands in association with the . Explain why Okazaki fragments are formed. Replication process. We demonstrate that the nonhydrolyzable ATP analog, ATPγS, supports the formation of an isolable leading strand complex that loads and replicates the lagging strand only in the presence of ATP, β, and the single-stranded . To deal with this problem, DNA synthesis is carried out discontinuously in a 5' to 3' direction. Describe the process of DNA replication and the functions of the enzymes involved. DNA polymerase also proofreads each new DNA strand to make sure that there are no errors.Primase synthesizes an RNA primer.An associated 3′–5′ exonuclease activity allows a polymerase to remove misincorporated nucleotides, and ensures the high-fidelity DNA synthesis that is required for faithful replication .The primase generates short strands of RNA that bind to the single-stranded DNA to initiate DNA synthesis by the DNA polymerase.Mécanisme Démarrage. D) DNA helicase.
DNA structure and replication review (article)
Pre-replication complex. On the leading strand, DNA is synthesized continuously, whereas on the lagging strand, DNA is .Three more DNA polymerases have been found in E.In contrast, polymerase δ (delta) synthesizes DNA on the opposite template strand in a fragmented, or discontinuous, manner and this strand is termed the “lagging strand” .
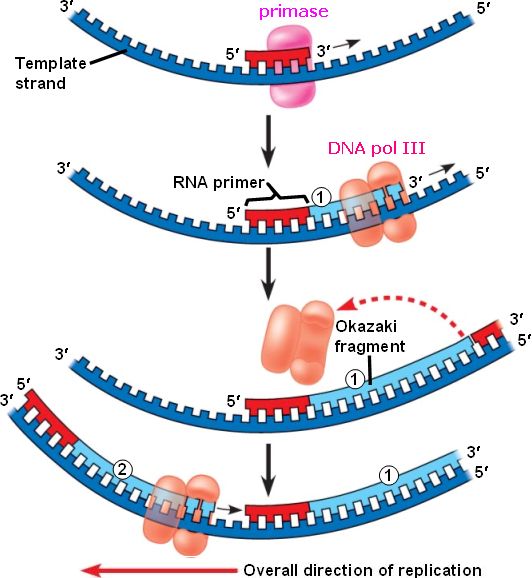
The enzyme DNA primase, which is present close to the opening of the replication fork, will synthesize multiple RNA primers on the lagging strand as the DNA unwinds.The DNA polymerase III holoenzyme is composed of 10 subunits. The core of the polymerase .DNA polymerase III then binds to the RNA primers and initiates DNA synthesis, synthesizing short Okazaki fragments in the 5′ to 3′ direction away from the replication fork.
Toutes les ADN polymérases synthétisent l’ADN dans le sens 5’ → 3’, chez tous les organismes vivants, et aucune n’est capable de commencer la synthèse .DNA polymerase I (pol I) processes RNA primers during lagging-strand synthesis and fills small gaps during DNA repair reactions. Genes (Basel) . A replication .DNA polymerases are the enzymes that build DNA in cells. Each Okazaki fragment ranges from 100 to 1000 nucleotides in length.DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae - PMC.
Leading and lagging strands in DNA replication
Imagine that you have recreated such a life form.
Leading Strand and Lagging Strand Synthesis
The leading strand is built .
USA 111, 15390–15395 . It is possible that the flexible linker connecting the motor domains of τ with the carboxy-terminal protein binding sequences in τ provides a range .
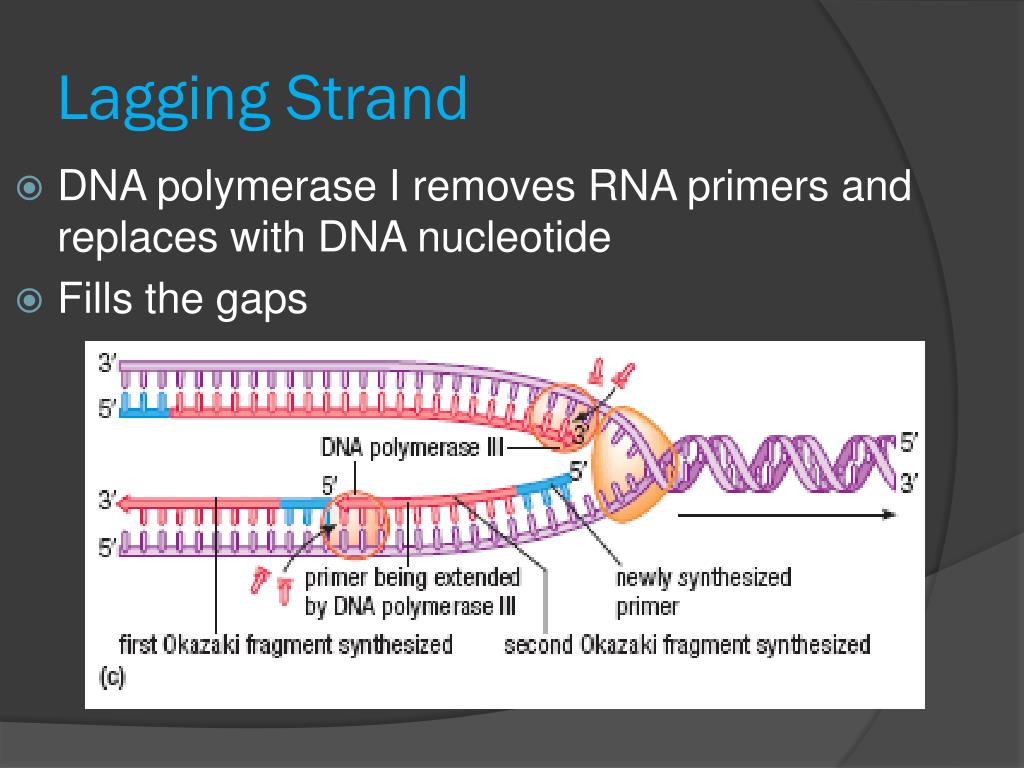
The lagging strand is synthesized discontinuously in short Okazaki fragments, each requiring its own primerHowever, DNA polymerase cannot synthesize DNA in a 3' to 5' direction on the lagging strand. During elongation, the leading strand of DNA is synthesized continuously from a single primer.At this point, DNA polymerase III releases the lagging strand and its place is taken by DNA polymerase I, which does have a 5′→3′ exonuclease and so removes the primer, and usually the start of the DNA component of the Okazaki fragment as well, extending the 3′ end of the adjacent fragment into the region of the template that is exposed. Once initiated, a replication bubble (replicon) forms as repeated cycles of elongation proceed at opposite replication forks. Leading strand.Explain why DNA replication is bidirectional and includes both a leading and lagging strand. In bacteria, DNA polymerase III binds to the 3′-OH group of the nicked strand and begins to unidirectionally replicate the DNA using the un-nicked strand as a template, displacing the nicked strand as it does so.Duplex DNA is antiparallel, and therefore even though the lagging strand polymerase moves with the replication apparatus, it synthesises the lagging strand in the direction opposite fork progression. It was discovered by Thomas Kornberg (son of Arthur Kornberg) and .The DNA Polymerase III holoenzyme forms initiation complexes on primed DNA in an ATP-dependent reaction. The core of the polymerase contains the catalytic polymerase subunit, α, the proofreading 3′ → 5′ exonuclease, ε, and a subunit of unknown function, . Some scientists have proposed that the earliest forms of life may have existed in an RNA World where RNA was both the genetic material and responsible for enzymatic activity.DNA polymerase.As it is well established that 3′-terminal mispairs are much more difficult to extend than correct base pairs (27, 28, 35 –37), and this has been well demonstrated for E. Leading and lagging strands .
Analysis of mutational asymmetry with respect to the direction of replication and transcription suggests that error-prone damage bypass on the lagging strand has a major role in human germline and . Each strand then serves as a template for a new DNA molecule. One new strand, the leading . 10 Given the diagram above, what would be the correct labelling for numbers 1 and 2? 1: DNA Ligase; 2: Leading Strand 1: DNA polymerase III; 2: Leading strand 1: DNA polymerase III; 2: Single Stranded DNA Binding proteins 1: DNA polymerase I; 2: Lagging Strand 1: DNA polymerase I; 2: Single Stranded DNA . coli thioredoxin (trx).Most current evidence indicates that DNA polymerases ε and δ, respectively, perform the bulk of leading and lagging strand replication of the eukaryotic nuclear genome.At least three DNA polymerases are required for eukaryotic genome replication: DNA polymerase alpha (Pol α), DNA polymerase delta (Pol δ) and DNA .Because DNA polymerase can add nucleotides in only one direction (5' to 3'), the leading strand allows for continuous synthesis until the end of the chromosome is reached; however, as the replication complex arrives at the end of the lagging strand there is no place for the primase to land and synthesize an RNA primer so that the synthesis of the . E) DNA polymerase I.A primer is needed to start replication. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. A) DNA polymerase III. Leading strand is synthesised continuously. DNA polymerase with proofreading abilityAdditionally, RPA also influences lagging strand synthesis by stimulating the strand displacement activity of DNA polymerase d 16(pol d), which functions to create a . This process is called proofreading. Lagging strand.Here we have investigated this question in the bacterium Escherichia coli, in which the replicase (DNA polymerase III holoenzyme) contains two copies of the same . 179 Replication Initiation in E.Both enzymes are stimulated by PCNA, although to different levels.DNA replication occurs in both directions. DNA is made differently on the two strands at a replication fork.Describe how DNA is replicated in eukaryotes. The discontinuous synthesis of the lagging strand necessitates the periodic synthesis of .
Molecular Events of DNA Replication
coli DNA polymerase III (10, 21, 36, 37), it is reasonable to assume that delays in lagging-strand synthesis at polymerase errors may lead to dissociation from the mismatch. Recalling that new nucleotides can only be added to the free 3' hydroxyl group of a pre-existing nucleic acid strand. Function DNA polymerase moves along the old strand in the 3'–5' direction, creating a new strand having a 5'–3' direction.Positional enrichment: whereas at OP sites the frequency of leading-strand replication by pol I will be <100% (as it is partially replaced by pol III) and will vary depending on distance from ori, the frequency of pol I lagging-strand replication is 100%, because all Okazaki primers need to be processed for successful replication. The leading strand can be extended by one primer alone, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. Preinitiation complex.Auteur : Sal Khan The overall direction of the lagging strand will be 3′ to 5′, and that of the leading strand 5′ to 3′.If the polymerase detects that a wrong (incorrectly paired) nucleotide has been added, it will remove and replace the nucleotide right away, before .DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication.The DNA polymerase is a 1:1 complex of the T7 gene 5 protein (gp5) and E.
DNA replication of the leading and lagging strand