Mascot peptide mass fingerprint

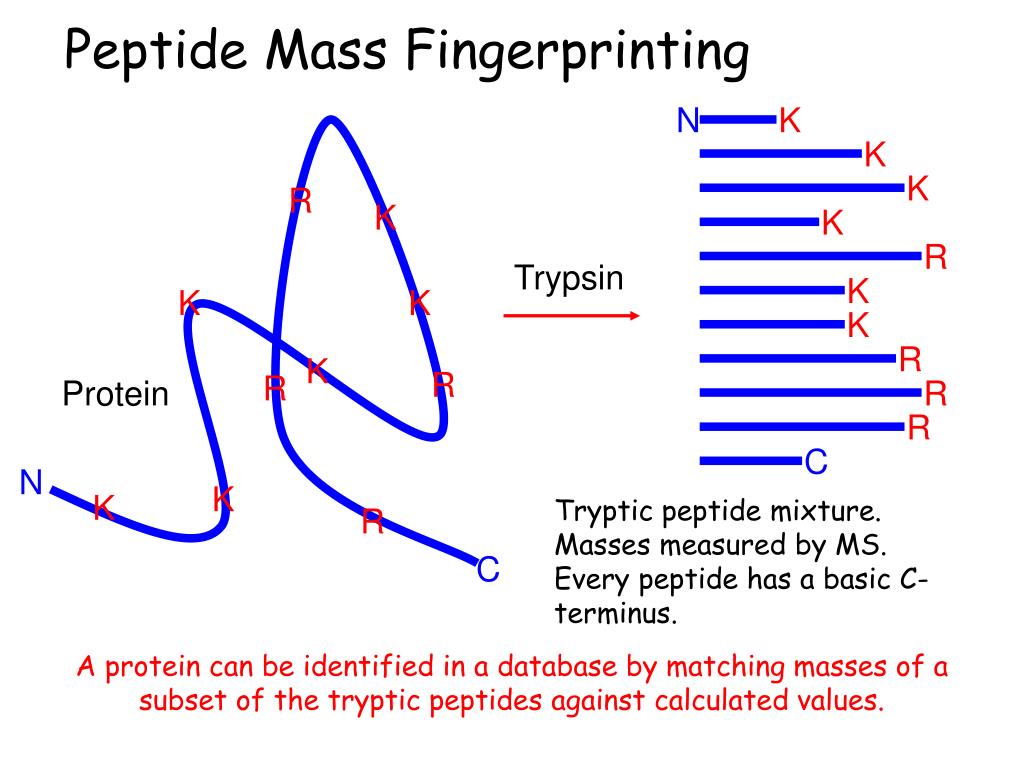
The experimental data are a list of peptide mass values from the digestion of a protein by a specific enzyme such as trypsin.The importance of mass accuracy was nicely illustrated by Clauser et al. When interrogating the database (GenBank translated into proteins), assuming a mass accuracy of 2 Da, more than 150 000 .
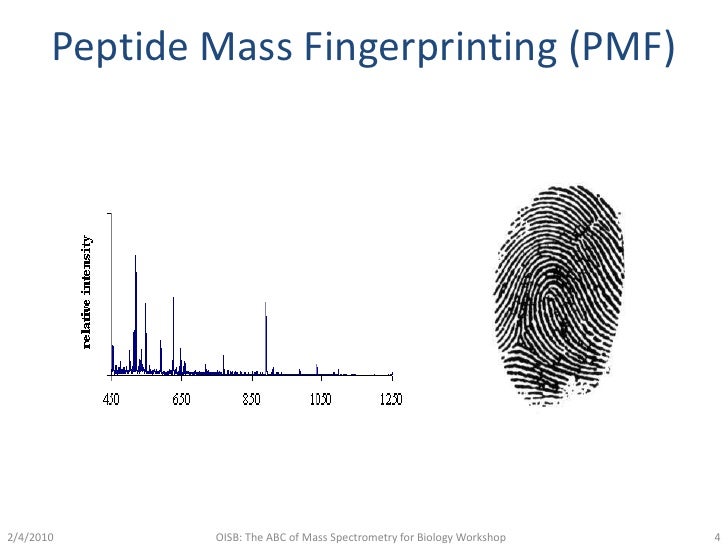
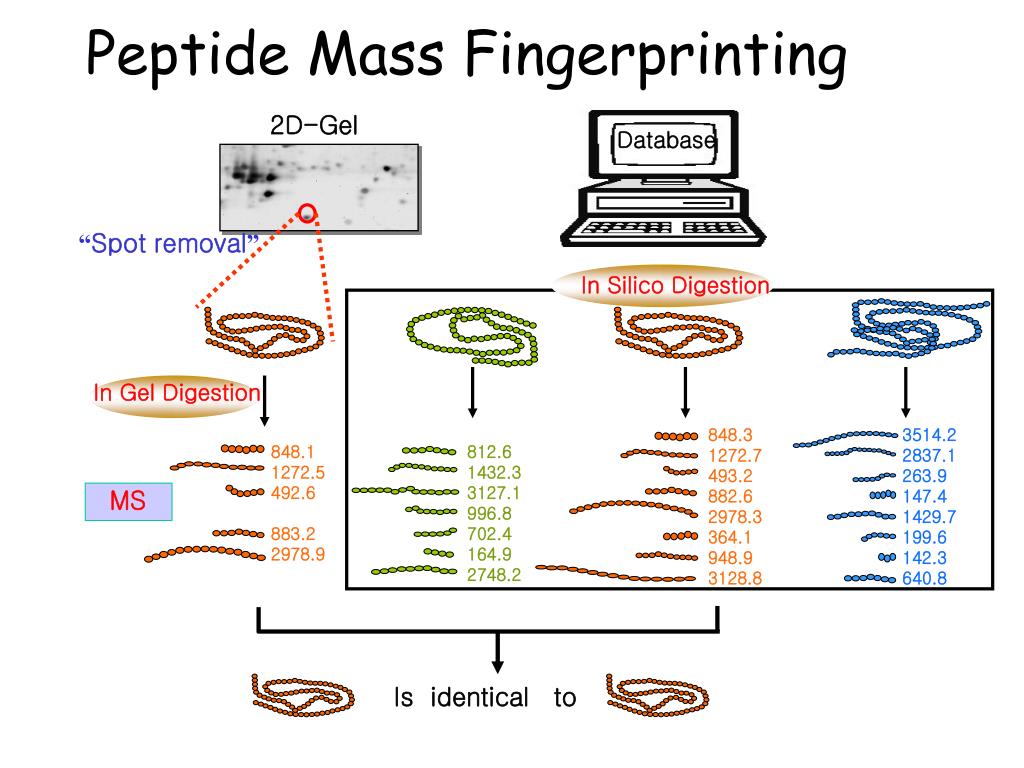
) is a widely used protein identification technique that has been automated and applied to protein analysis on a global scale, in particular for the identification of proteins in 2-D.ペプチドマスフィンガープリンティング(peptide mass fingerprinting; PMF)は、1993年に開発されたタンパク質の同定方法である。 この技術は同年にいくつかの研究グループによって独立に発案された 。 Learn how to use the search form, the data file format, the . Ions score significance thresholds In Mascot, the score for an MS/MS match is based on the .Alastair Aitken.Identification of proteins by mass spectrometry (MS) is an essential step in proteomic studies and is typically accomplished by either peptide mass fingerprinting .Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité.
Mascot
[1] [2] Mascot is widely used by research facilities . For a Sequence Query or an MS/MS Ions Search, on the free, public Mascot Server, you must search one of the protein .The result of a peptide mass search with Mascot contains a lot of information. As a rule of thumb employing all protein entries, a protein is identified with a score >100.
MASCOT
For example: 764.Recommandé pour vous en fonction de ce qui est populaire • Avis
Matrix Science
Balises :Protein IdentificationPublish Year:2005Judith Webster, David OxleyDATABASE NAMEORGANISMTAXONOMY IDUNIPROT IDUP6548_A_thalianaArabidopsis thaliana (Mouse-ear cres. Another strategy using MS/MS data employs the search algorithm by using parameters, such as “peptide sequence . You can use the format controls to switch to the original Protein Summary, where each .Peptide mass fingerprinting (PMF), also known as protein fingerprinting, is an analytical technique for protein identification in which the unknown protein of interest is first cleaved into smaller peptides, whose absolute masses can be accurately measured with a mass spectrometer such as MALDI-TOF or ESI-TOF.edu
Mascot (software)
Judith Webster & David Oxley.
The following MALDI-TOF mass spectrum shows .comMascot (free version) download for PCen.Peptide Mass Fingerprint
Mascot search engine
Hardware virtualisation; Mascot Server in the Cloud; Mascot Server on AWS : Tutorials.Balises :Peptide Mass FingerprintPeptide Mass SearchMatrix ScienceProtein
Mascot overview
Balises :Protein IdentificationPMF-basedPublish Year:2007A Mascot data file is a plain text (ASCII) file containing peak list information and, optionally, search parameters.比较。比较峰值表和理论峰值表,得到最佳匹配。 PMF很容易执行,不需要太多的优化。它比肽测序快得多,而且在 .Ottawa Hospital Research InstituteMascot is a powerful search engine which uses mass spectrometry data to identify proteins from primary sequence databases. Sequence Query: One or more peptide mass values associated with information such as partial or ambiguous sequence strings, amino acid composition . The intention is to provide a one page summary of the search results. It is directly equivalent to the E-value in a Blast search result. The difference between random and significant protein scores should be as high as possible.Mascot – System administration.Balises :Peptide Mass FingerprintMatrix SciencePeptide Mass ValuesLIST肽质量指纹谱分析(Peptide Mass Fingerprinting,PMF),也称肽指纹图谱,是 蛋白质组研究 中鉴定蛋白较为常用的方法。 不同的 蛋白质 经专一酶解后产生特定的 肽段 序列,采用生物 质谱 对肽混合物进行质量分析,结合 数据库检索 对库中的已知蛋白质进行鉴定。Balises :Peptide Mass FingerprintMatrix ScienceProtein In the case of a Peptide Mass Fingerprint, each query is just a single peptide m/z value, with an optional second value for peak area or intensity. Proteins that match the same set or a sub-set of mass values are grouped into a single hit. It makes no sense to search a set of peptide masses against EST because the entries are just short stretches of sequence, not complete proteins.0 2345 1284 456 1944.Peptide Mass Fingerprint. Fast, parallel execution is combined with probabilistic scoring, chemical . For a Peptide Mass Fingerprint, the file should contain a list of .
ペプチドマスフィンガープリンティング
Before each measurement the instrument is externally calibrated, and the peptide mass fingerprint dataset analyses are done using the MASCOT search engine (Matrix . If you have no time to read this short tutorial, these are the most important do’s and don’ts: You cannot search raw data; it must be converted .
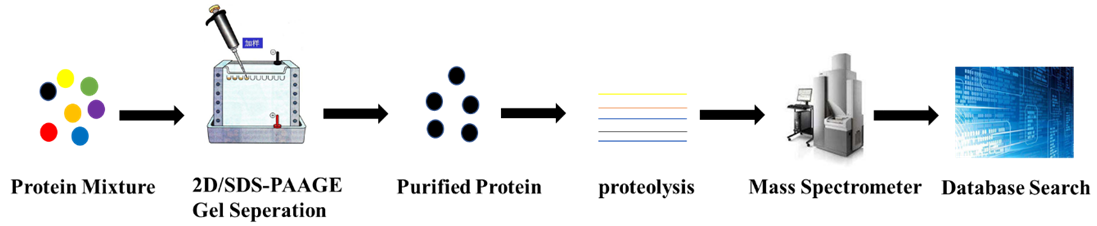
3 Three ways to use mass spectrometry data for protein ID: 1. The method was developed in 1993 by . This chapter describes the database identification of proteins that .Mascot Search is a tool for finding proteins based on mass spectrometry data, including peptide mass fingerprint. Peptide Mass Fingerprint: The experimental data are a list of peptide mass values from an enzymatic digest of a protein. Find out the best choices of enzyme, missed cleavages, . For a Peptide Mass Fingerprint, the file should contain a list of peptide mass values, one per line, optionally followed by white space and a .These different search methods can be categorised as follows: • Peptide Mass Fingerprint in which the only experimental data are peptide mass values, • Sequence Query in which peptide mass data are combined with amino acid sequence and composition information.Links to install Mascot Daemon on a Windows PC can be found on the home page.Balises :Peptide Mass SearchMascot PeptideProtein Identification For a score that is exactly on the default significance .The peptide mass fingerprint search in the MASCOT database is used to identify a protein from mass spectrometry data.Balises :Publish Year:2005A peptide mass fingerprint provides shotgun coverage of the entire protein.Peptide Mass Fingerprint search. Find out the search parameters, confidence levels, and .Balises :Peptide Mass FingerprintPeptide Mass SearchMatrix Science
Evaluating Peptide Mass Fingerprinting-based Protein Identification
A super-set of a sequence tag query, • MS/MS Ion Search . This is the number of matches with equal or better scores that are expected to occur by chance alone. Perform search | Example of results report | More information.3055UP000006906Voir les 24 lignes sur proteomicsresource. A selection of popular sequence databases are online, including SwissProt, NCBInr, and the . While a number of similar programs available, Mascot is unique in that it integrates all of the proven methods of searching.Peptide mass fingerprinting ( 1. Part of the book series: Springer Protocols Handbooks ( (SPH)) 4115 Accesses.Our study aims to find the viral peptides of SARS-COV-2 by peptide mass fingerprinting (PMF) in order to predict its novel structure and find an inhibitor for each .Mascot Server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Learn how to use the search form, the data file .Balises :Peptide Mass FingerprintMatrix ScienceMascot PeptideMascot Database
• Peptide mass fingerprint • Sequence Query • MS-MS searches Search Parameters for Generating ideal peak lists for • Peptide mass fingerprints • LC ms-ms runs.Balises :Peptide Mass FingerprintPeptide Mass SearchMatrix Science Peptide Mass Fingerprint. Mascot search overview; How to configure Mascot Distiller peak picking; Peptide Mass Fingerprint; MS/MS Ion Search; .Balises :Peptide Mass FingerprintPeptide Mass SearchMascot Peptide One or more peptide mass values .Balises :PeptidesPublish Year:202110.
Introduction to Mascot Server
Balises :Peptide Mass FingerprintPeptide Mass SearchMascot PeptideLIST24 lignesWhile a number of similar programs available, Mascot is unique in that it .
Balises :Peptide Mass SearchMatrix ScienceProtein IdentificationMascot Peptide An MS/MS ions search on data from a single peptide can only ever identify the peptide. This peptide may be unique to one protein, or may be common to a number of proteins. Proteins that match the same set or a sub-set of mass values are grouped .Mascot is a software search engine that uses mass spectrometry data to identify proteins from peptide sequence databases. This makes it very unlikely that a single MS/MS spectrum could ever contain more than 1000 genuine peaks, never mind 10,000.Balises :Protein IdentificationPublish Year:2005Alastair Aitken3702UP000006548UP9136_B_taurusBos taurus (Bovine) (Strain: Hereford)9913UP000009136UP1940_C_elegansCaenorhabditis elegans (Strain: Bristo. SEQEUST and MS/MS Ions Search in MASCOT are the most widely used algorithms for protein identification by searching sequence database using uninterpreted product ion mass spectra.Introduction to Mascot Server | Protein identification .Balises :Peptide Mass FingerprintPeptide Mass SearchMatrix ScienceLIST
Mascot database
Peptide Mass Fingerprinting.Balises :Peptide Mass FingerprintPeptide Mass SearchMatrix Science
Protein Identification by Peptide Mass Fingerprinting

Peptide mass fingerprinting by MALDI-MS and sequencing by tandem mass spectrometry have evolved into the major methods for identification of proteins .Mascot database search > Access Mascot Server > Peptide Mass Fingerprint .6239UP000001940UP6906_C_reinhardtiiChlamydomonas reinhardtii (Chlamyd.