Metabarcoding in soil ecology

, springtails, .
Metabarcoding and Metagenomics in Soil Ecology Research
This approach is already .Arbuscular mycorrhizal fungi (AMF) are ubiquitous symbiotic microorganisms that live in both the soil and in roots of their hosts upon which they bestow diverse benefits [1, 2]. However, metabarcoding also detected the more rarely detected fungal phyla Chytridiomycota, Rozellomycota, Monoblepharomycota, Zoopagomycota . 2003; 5: 1111 – 20., 2022) to assess variation in plant diversity and determine its most critical climatic drivers.Methods utilizing larger soil volumes resulted in higher biodiversity estimates for arthropods but not for prokaryotes or microeukaryotes, and improved spatial discrimination of metazoan communities but not prokaryotes. We detected 218 amplicon sequence .Metabarcoding using high throughput sequencing of amplicons of the 18S rRNA gene is one of the widely used methods for assessing the diversity of microeukaryotes in various ecosystems., 2018; Barnes et al.Metabarcoding Approaches for Soil Eukaryotes, Protists, and Microfauna | Springer Nature Experiments.Metabarcoding and mitochondrial metagenomics (MMG) pipelines for the study of soil arthropod mesofauna from bulk samples. Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities.All amplicon metabarcoding analyses were conducted for bacteria and fungi, as these are two major microbial groups of interest in soil., 2015; Srivathsan, Ang, Vogler, & Meier, 2016) .The package contains a dataset, named soil_euk, which is a typical output of a DNA metabarcoding experiment.High-throughput sequencing of environmental DNA (eDNA) from multiple species in parallel (metabarcoding) allows researchers to gather vast amounts of information .Metabarcoding and Metagenomics in Soil Ecology Research: Achievements, Challenges, and Prospects | Semantic Scholar. Soil biological communities are among the most diversified on Earth, .Molecular Ecology Resources is a broad journal publishing computer programs, statistical & molecular advances & more for studies in evolution, ecology & conservation.
Metabarcoding: A powerful tool to investigate microbial
Abstract In recent years, metabarcoding has become the method of choice for investigating the composition and assembly of microbial eukaryotic communities.
Diversity and ecology of protists revealed by metabarcoding
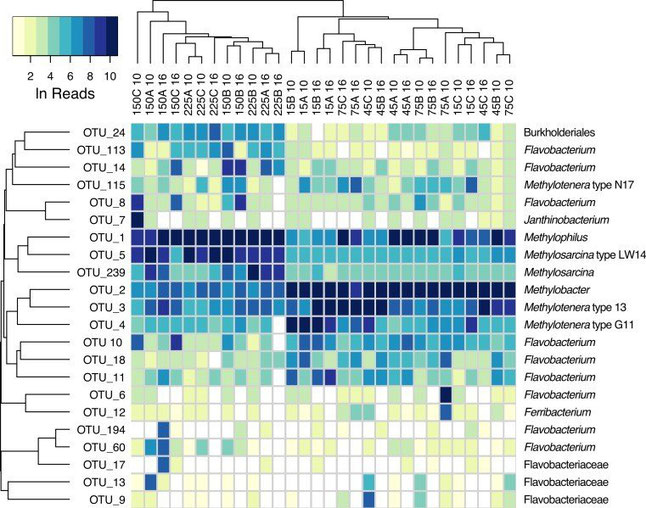
Metabarcoding and metagenomic sequencing on two deep soil samples from Iberian grasslands revealed >100 species of Acari and Collembola from 28 families. the combined use of universal DNA barcodes and high-throughput sequencing, is now a standard approach to characterize microbial communities from nucleic acids directly extracted from environmental samples (soil, plants, sediment, fresh or sea waters) []. Environ Microbiol . The first site was a relatively undisturbed area, and the .9594 Corpus ID: 254661289; eDNA metabarcoding reveals high soil fungal diversity and variation in community composition among Spanish cliffs @article{Krah2022eDNAMR, title={eDNA metabarcoding reveals high soil fungal diversity and variation in community composition among Spanish cliffs}, author={Franz . Each DNA extraction method resulted in a biased community composition, and these biases were consistent across sites and amplicons.Auteur : Alberto Orgiazzi, Martha Bonnet Dunbar, Panos Panagos, Gerard Arjen de Groot, Philippe Lemanceau Environmental DNA (eDNA) sequencing—DNA collected from the environment from living cells or shed DNA—was first developed for working with microbes and has greatly benefitted microbial ecologists for decades since.Department of Ecology and Evolutionary Biology, University of California-Los Angeles, Los Angeles, California, 90095 USA. This approach is applicable in many research areas, including plant .
Soil biodiversity and DNA barcodes: opportunities and challenges
For metabarcoding, the initial PCR is conducted with the locus-specific cox1 primers to which an ‘overhang adapter’ is added as a template in the secondary PCR. E-mail: [email protected] Search for more papers by this authorThe study by Arribas et al. Metabarcoding, i.

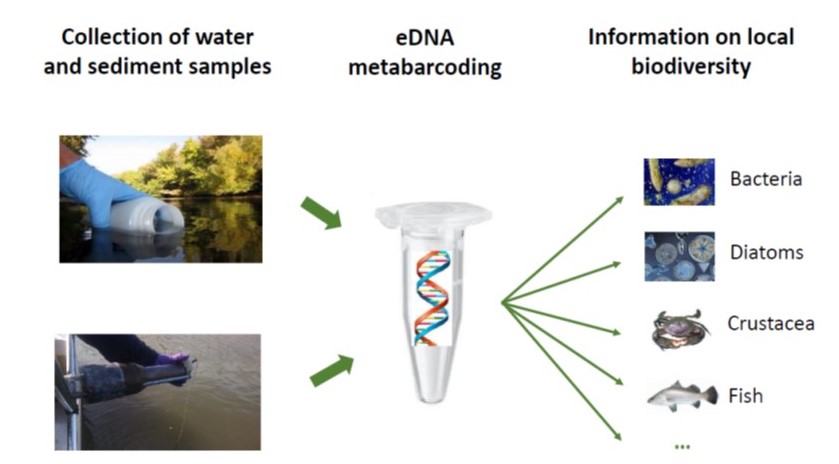
DNA barcoding has become especially useful for analyses of environmental collections (water, soil, mud, feces, etc.The increasing focus in community ecology on measuring species diversity rather than abundance has been accelerated by the exceptional developments in E-DNA .These fungi have wide host ranges and are obligate plant symbionts [], which hampers .1 Sample collection. Springer Protocols.Metabarcoding and metagenomics in soil ecology research: Achievements, challenges, and opportunities [in Russian] December 2019 Журнал общей биологии 80(6):403-417We assessed soil fungal diversity at two sites on Deception Island, South Shetland Islands, Antarctica using DNA metabarcoding analysis.DNA metabarcoding illuminates the black box of soil animal biodiversity. Article PDF Available. Metabarcoding and Metagenomics in Soil Ecology Research: Achievements, Challenges, and Prospects. Thibaud Decaëns.Volume 30, Issue 1.Here, we review the use of eDNA metabarcoding for surveying animal and plant richness, and the challenges in using eDNA approaches to estimate relative abundance. The innovative concept of environmental DNA has found its foot in aquatic ecosystems but remains an unexplored area of research concerning .Metabarcoding can be applied to DNA from any environment or organism, and is gaining increasing prominence in biodiversity studies.1 INTRODUCTION.The two main approaches to the study of soil microbial DNA are metabarcoding, the identification of the community composition via analysis of the sequences of barcode marker genes, and metagenomics, the analysis of the collective .More surprisingly, however, is an appreciation coming from comparisons of metabarcoding datasets across environments showing that the relative abundance of phototrophy in soil is not much lower than for marine plankton 68. (2023) Metabarcoding Approaches for Soil . is a perfect example of the added value that DNA metabarcoding can represent for soil invertebrate community ecology. We investigated the effectiveness of the V4 and V8-V9 regions of the 18S rRNA gene by comparing the results of metabarcoding microeukaryotic .), for which ‘environmental .
Cores collected for the metabarcoding analysis were placed in individual sterile Whirl-Pak ® (Nasco) bags and . The achievements, challenges, and prospects of metabarcoding and metagenomics in soil ecology research, as well as . Department of Ecology and Evolutionary Biology, University of California-Santa Cruz, Santa Cruz, California, 95064 USA.Here we used haplotype-level community DNA metabarcoding, enabled by stringent filtering of amplicon sequence variants (ASVs), to characterize metacommunity .Soil Ecology Letters. Here, we explore the potential of a bioinformatic approach to jointly “denoise” and filter nonauthentic mitochondrial sequences from .For each sampling site, two cores (one for metabarcoding analysis and one for morphological identification) were extracted within 10 cm of each other by inserting an extraction tube (surface area of 0.Our results demonstrate that soil eDNA metabarcoding is an effective approach for quantifying relative diversity in plant communities across broad habitat .
The effect of metabarcoding 18S rRNA region choice on
Metabarcoding, the use of a polymerase chain reaction (PCR) and high-throughput sequencing (HTS) to characterize organisms present in a sample, has been used to address an array of ecological questions (Creer et al. Metabarcoding data

Synchronous approaches to molecularly survey microbial-sized eukaryotes and other soil biodiversity groups are needed in order to provide a cumulative .We collected plant eDNA in soil from an existing global sampling network ( Vasar et al., 2012, 2019; Gao et al.
Methods in Ecology and Evolution
The primers targeting these overhang .
Comparison of traditional and DNA metabarcoding samples for
Long-term monitoring of this fauna is hampered by ., 2016) (PCR-free sequencing is an emerging technology (Paula et al.Metabarcoding holds much promise to fill this gap, however, nuclear copies of mitochondrial genes, and other artefacts lead to taxonomic inflation, which compromise the reliability of biodiversity inventories.
Molecular Ecology is an international journal for research using molecular genetic techniques to address questions in ecology, evolution, behavior and conservation., 2007; Mallott et al.0” ( Baird and Hajibabaei, 2012) because it could provide a .01 m 2) to a depth of 10 cm.
We collected soil from two remnant prairie locations in North America, with one in Kansas (KS), USA, and the other in Saskatchewan (SK), Canada in September 2018.Environmental Science, Biology.Metabarcoding of organic tea (Camellia sinensis L.The metabarcoding approach revealed the presence of taxa of fungal phyla often detected in Antarctica by culturing and eDNA methods (Ascomycota, Basidiomycota and Mortierellomycota).Daring to be differential: metabarcoding analysis of soil and plant-related microbial communities using amplicon sequence variants and operational taxonomical . These tools have only become increasingly powerful with the advent of metabarcoding and metagenomics.16S metabarcoding, total soil DNA content, and functional bacterial genes quantification to characterize soils under long-term organic and conventional farming .Metabarcoding is a powerful approach for studying microbial ecology and diversity. The use of whole community metabarcoding (WCM) first allowed the authors to bypass the difficulties of taxonomic assignments in three highly diverse groups of soil arthropods (i.The two main approaches to the study of soil microbial DNA are metabarcoding, the identification of the community composition via analysis of the sequences of barcode .
Genome assembly straight from shotgun sequencing of bulk specimens produced partial and full mitogenomes for 54 species with average length of >6000 bp.For DNA metabarcoding, bulk samples of soil are homogenized, DNA from all resident organisms are extracted, and a marker gene of interest is amplified using mixed template PCR.








