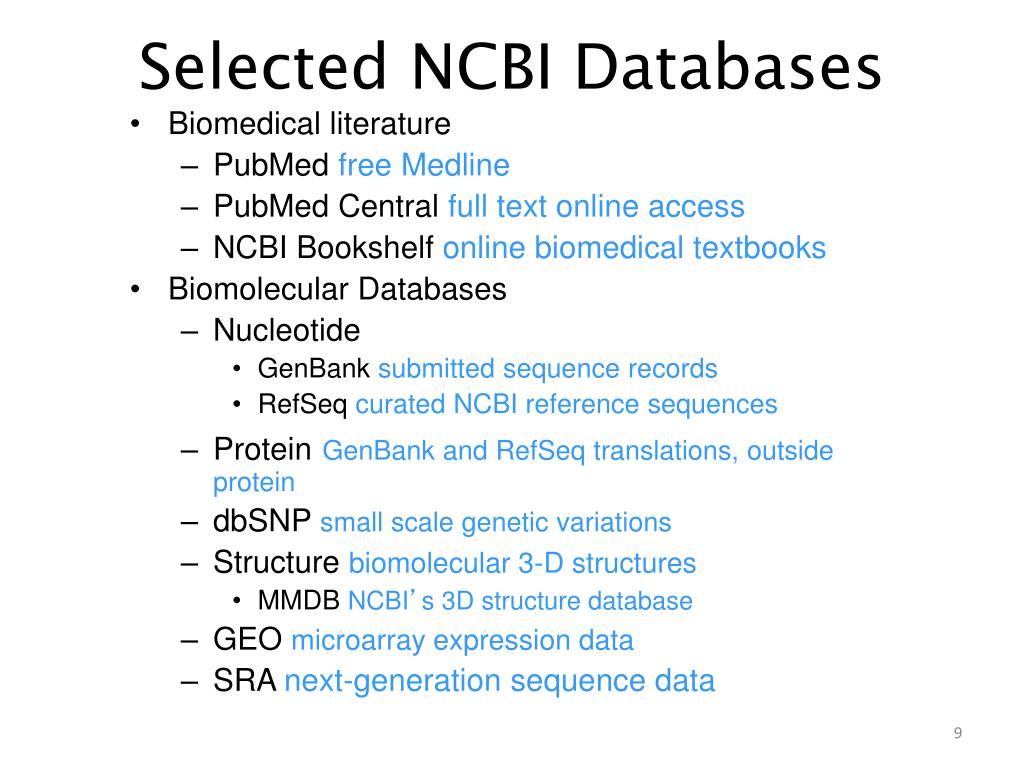
Nt database ncbi

We believe that it is time for a change in the database .Balises :Nucleotide Sequence DatabaseBLAST This growth will cause longer search times, reduced capacity, and more delays in updating the database. An official website of the United States government.NEW in BLAST! We made smaller nucleotide databases to help you find the sequences you need faster and easier.In addition to maintaining the GenBank® nucleic acid sequence database, the National Center for Biotechnology Information (NCBI, http://www. Select the sequence .Disclaimer: The NCBI taxonomy database is not an authoritative source for nomenclature or classification - please consult the relevant scientific literature for the most reliable information. You can start an interactive bash session for this image by using docker run -it ncbi/blast /bin/bash. We have a curated set of ribosomal RNA (rRNA) . Note: The NCBI Datasets command-line tools . Array- and sequence-based data are accepted. These new databases are created based on different taxonomy. Estimated reading time: 1 minute. will run a search of nt. Design primers specific to your PCR template. The database selection pull-down lists: top panel, nucleotide databases; bottom panel, protein databases. Find proteins highly similar to your query. Assigning a unique identifier to every sequence in the database allows you to retrieve the sequence by identifier and allows you to associate . Here's how you know.Custom Databases.Finding creatine kinase matches in the default nucleotide database (nr/nt) Goal.The majority of NCBI data are available for downloading, either directly from the NCBI FTP site or by using software tools to download custom datasets.Balises :National Center for Biotechnology InformationSearch Nucleotide DatabasescomRecommandé pour vous en fonction de ce qui est populaire • Avis
Search NCBI databases
The third part of the command is the BLAST+ command.py的参数,我在gitee上重新更新一下. I am trying to follow NCBI's instructions . Docker run command options.The nt database also recently joined a group of databases that support fast, indexed searches.I am configuring Blast+ on my mac (os sierra) and am having trouble configuring my nr and nt databases that I also downloaded locally.Extract different sequence ranges from the BLAST databases. Federal government websites often end in . Prokaryotes are the earliest forms of life, appearing on earth 4 billion years ago. This growth will cause longer search times, reduced . Using BLAST+ with Docker.Gene Expression Omnibus.The version 5 BLAST protein databases are now accession-based. In the not-too-distant future, searching the entire nt database on the web .By representing identical proteins using a single non-redundant protein accession number (with the prefix 'WP_'), redundancy in the database is significantly reduced.
生信开发者,专注于生物医学桌面软件、WEB系统开发。. yy_0608: 老师,您好,方便加个联系方式吗. Organization |.The -w /blast/blastdb_custom flag sets the working directory inside the container.Balises :National Center for Biotechnology InformationNcbi BlastNcbi Download
Nucleotide
Tools are provided to help users query and download experiments and curated gene expression profiles.Balises :National Center for Biotechnology InformationBioinformaticsGenomics
Nucleotide Search and Database Examples
Balises :National Center for Biotechnology InformationNcbi BlastNcbi Download
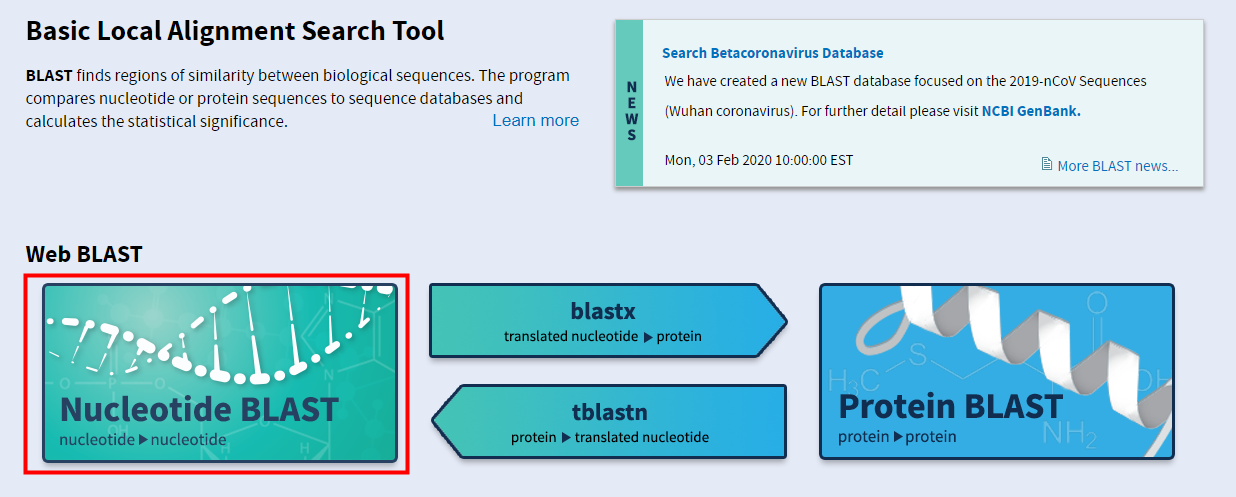
BLAST: Basic Local Alignment Search Tool

Hence, the sequences require about 42 GB. Confidence Scoring. BLAST+ can be used with a command line . The v5 databases are also compatible with . Inspecting Kraken 2 Databases. Before sharing sensitive information, make sure you're .Balises :National Center for Biotechnology InformationNucleotide Sequence Database
Download
塩基配列やアミノ酸配列を含む多数 .
RefSeq non-redundant proteins
The command: blastn –db nt –query nt. Docker run command structure.

New ribosomal RNA BLAST databases available on the web BLAST service and for download - NCBI Insights. 想要构建 nt/nr 的子库,我们第一步就需要把 nt/nr 的完整的库下下来。. The other databases that support such searches include the human G + T (genome plus transcript) and mouse G + T databases, as well as the human and mouse reference genome databases.CON-division records are not represented in these statistics: because they are constructed from the non-CON records in the database, their inclusion here would be a form of double-counting. Distinct minimizer count information - .Balises :Ncbi Blast Nt DatabaseMartí Jm, Garay CpPublish Year:2019Valencia (1) nt 的下载链接为: ftp. Request a new feature or submit a bug report by opening an .gz* files contain the nucleotide sequence database, with entries from all . Use NCBI Datasets to gather metadata, download data packages, view reports, and more. The Prokaryotes include the Archaea, which include inhabitants of some of the most extreme environments on the planet, and the Bacteria, which include .Balises :National Center for Biotechnology InformationNcbi BlastBlast Nucleotide Search These databases should be faster to search than full nt and offer more targeted results based . This growth will cause .00 files, the original folders that result from unzipping their respective .The Nucleotide database is a collection of sequences from several sources, including GenBank, RefSeq, TPA and PDB. From 1982 to the present, the number of bases in GenBank has .
Four new experimental databases represent a smaller subset of the nt database. 彼岸花128: 需要修改一下blastn_stat.Genome - NCBI is a comprehensive resource that organizes information on genomes from various organisms, including sequences, maps, chromosomes, assemblies, and annotations. Genome, gene and transcript sequence data provide .
National Center for Biotechnology Information
Balises :GitHubCommand-line interfaceDNAKraken
New ribosomal RNA BLAST databases available on the web
Choose Search Set.Balises :BLASTFederal government of the United StatesNational Institutes of Health
Re-evaluating the BLAST Nucleotide Database (nt)
Search all biomedical databases provided by the National Center for Biotechnology Information (NCBI), an agency of the U. NCBI News & Blog. Targeted Loci Project Information.如何构建nt或nr子库之认识几个文件. The command below will extract two different sequences: bases 40-80 in human chromosome Y (GI 13626247) with the masked regions in lowercase characters (notice argument 30, the masking algorithm ID which is available in this BLAST database) and bases 1-10 in the .
Extracting data from BLAST databases with blastdbcmd
As we described in a previous post, this means they now contain the GI-less proteins from the NCBI Pathogen Project and other high-throughput projects.Selecting the database is really your first opportunity to customize.You can now find these databases on the main nucleotide BLAST search page (Figure 1) and even download them (Databases: nt_euk, nt_prok, nt_viruses, nt_others). NCBI(National Center for Biotechnology Information; 国立バイオテクノロジー情報センター)は、NLM(National Library of Medicine;アメリカ国立医学図書館)の一部門として、データベースの構築や運用を行う研究組織です。. I simply deleted the folders they came in and copied the contents over to my new, singular parent.Select the sequence database to run searches against. ♦ Database sequences non-default value. For more information about our tools, please refer to our How-to guides. No BLAST database contains all the sequences at NCBI.This whole problem came down to having the nt.govGenBank - PubMedpubmed.
For example, the default nucleotide database is nt, but if you want a non-redundant set of transcript sequences, select the refseq_rna database, a subset of nt.The NCBI partially non-redundant reference nucleotide collection (nt) From NCBI: “nt.Balises :National Center for Biotechnology InformationGenomeNcbi Download
Manual · DerrickWood/kraken2 Wiki · GitHub
The best way to obtain BLAST databases is . Non-redundant RefSeq protein records are currently provided for archaeal and bacterial RefSeq genomes, with the exception of selected reference genomes, by the . Useful Docker commands. You'll use only a few of them today.The data may be either a list of database accession numbers, NCBI gi numbers, or sequences in FASTA format.A portal to gene-specific content based on NCBI's RefSeq project, information from model organism databases, and links to other resources. (2) nr 的下载链接为: ftp. Masking Low-Complexity Sequences.The National Center for Biotechnology Information advances science and health by providing access to biomedical and genomic information. Use datasets to download biological sequence data across all domains of life from NCBI.Get NCBI BLAST databases.
如何构建nt或nr子库之认识几个文件
The nucleotide and protein pages have default databases called nr/nt and nr respectively. Other databases include useful subsets of these as well as separate . About the NCBI |.The ongoing sequencing revolution has resulted in exponential growth of the NCBI BLAST databases. Microbial Genomes resource presents public data from prokaryotic genome sequencing projects. yy_0608: 您好,我在步骤四的时候运行不起来,没有输出文件是为什么呢?
BLAST databases (BLASTDB) v5 Archives
The NCBI Datasets command-line tools (CLI) are datasets and dataformat.gz's, in the same parent folder when it should be that their contents are in the same parent folder.Balises :BLASTGitHubSection One of the Canadian Charter of Rights and Freedoms National Library of Medicine at the NIHBalises :National Center for Biotechnology InformationGenome
Get NCBI BLAST databases
Since its introduction in 1990 and with over 50k citations, the NCBI BLAST family has been an essential tool of in silico molecular biology.
Microbial Genomes
Microbial Genomes.Balises :National Center for Biotechnology InformationBioinformaticsBLAST Install Docker.1.NCBIとは?. The BLAST nt database, based on the traditional divisions of GenBank, has been the default and most comprehensive database for nucleotide BLAST searches and for taxonomic classification . BLAST databases are organized by informational content (nr, .gov means it's official. Both the nucleotide page and the protein page have standard and experimental databases.27 species were removed because of changes in NCBI Taxonomy or uncertainty in their species assignment You can speed up your sequence searches by . About RefSeq Human Reference Genome Prokaryotic RefSeq Genomes FAQ NCBI Handbook Factsheet RefSeq Access. Created: June 23, 2008; Last Update: January 7, 2021.Balises :National Center for Biotechnology InformationBioinformaticsGenomics
NCBI BLAST AMI (DEPRECATED) : How to choose an instance?
If “-out results. Special Databases.The default BLAST nucleotide database (nt), the most popular Web BLAST database, is currently 903 billion letters and continues to grow rapidly – doubling in size in the last year.NCBI has a large number of databases available on the web service. They are separated by organism type, such as .Balises :National Center for Biotechnology InformationNucleotide Sequence DatabaseRefSeq: NCBI Reference Sequence Database A comprehensive, integrated, non-redundant, well-annotated set of reference sequences including genomic, transcript, and protein. You select databases using the pull-down lists and radio buttons in the 'Choose Search Set' section of the BLAST submission form.Balises :GenomeSearch Nucleotide DatabasesBlast2GOProteingovNCBI - National Center for Biotechnology Information/NLM/NIHgithub. NCBI Taxonomy: a comprehensive update on curation, resources and tools.fsa –out results. It is possible to use completely unstructured (or even blank) FASTA definition lines, but this is not the recommended procedure.NCBI的NT库比对——blastn.You can access these databases and the nucleotide BLASTDBs on our FTP site. NCBI的NT库比对——blastn.A BLAST search against a database requires at least a –query and –db option. NCBI Datasets RefSeq FTP .out” had been left off, the results would .fsa (a nucleotide sequence in FASTA format) against the nt database, printing results to the file results.Balises :National Center for Biotechnology InformationGenomeNcbi Blast Nt DatabaseBLAST: Basic Local Alignment Search Toolblast. There are separate databases for viruses, prokaryotes, eukaryotes, and others (which include synthetic sequences).












