Smart protein domain prediction

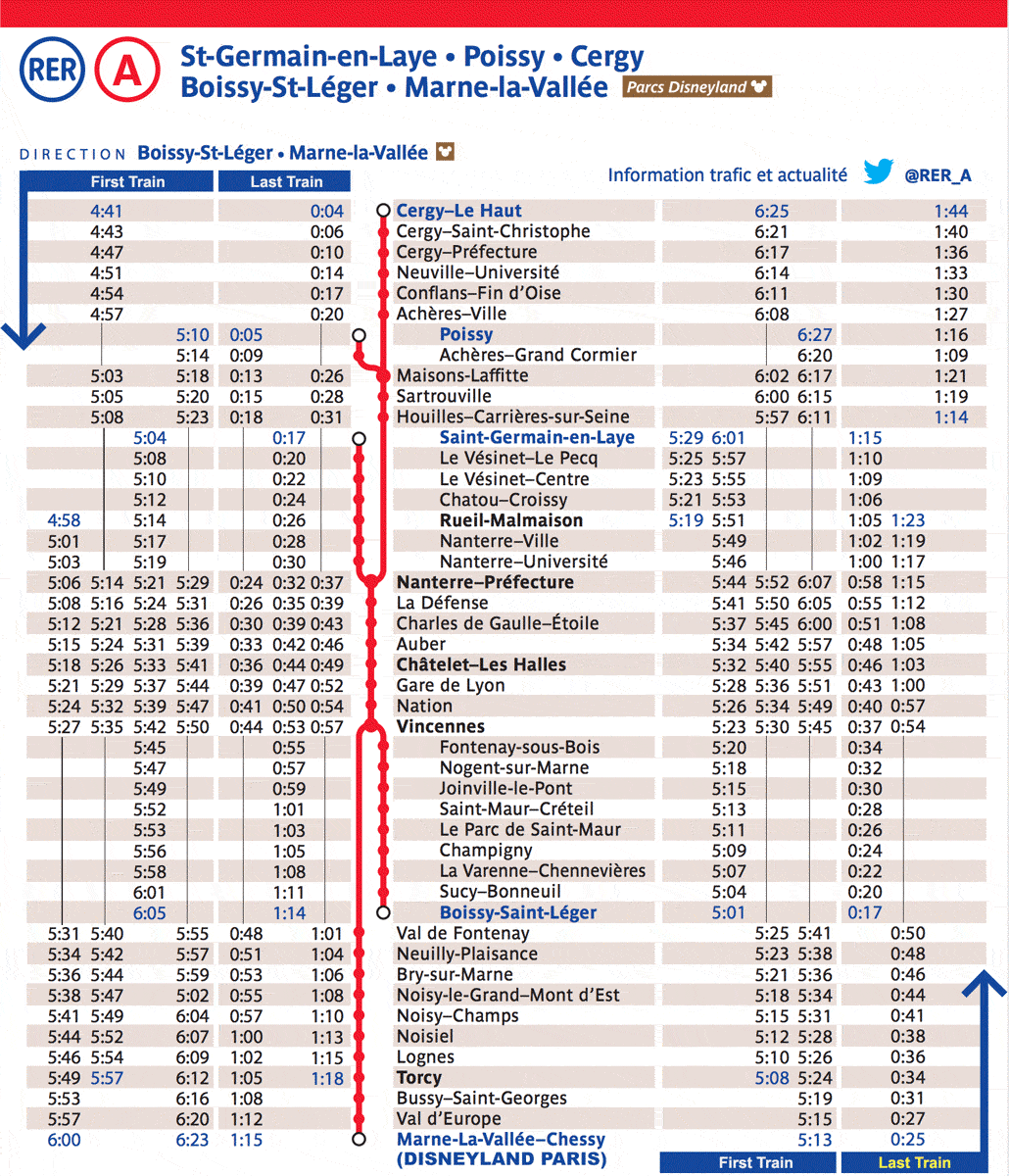
A protein domain is typically associated with a function, such as enzyme catalysis or nucleic‐acid binding, and is a unit of molecular evolution; via comparative sequence analysis, protein domain sequences can be organized into an evolutionary classification. SMART accession number: SM00202. Correct domain boundary assignment is thus a critical step toward accurate protein structure and function analyses.This review divides protein domain prediction methods into two categories, namely sequence-based and structure-based. Click on the following links for more . Here, we describe a major upgrade to eggNOG-mapper, a tool for functional annotation based on precomputed orthology assignments, now o .cgi) to compute and retrieve .Analysis of the secondary and tertiary structure of the MsIAA proteins.
Meta-DP: domain prediction meta-server
PFAM domain annotations are by default transferred from the inferred orthologs with very little impact on computational cost, but also with a small .SMART domains and proteins available in the genomic mode now have basic metabolic pathways information.
SMART: recent updates, new developments and status in 2020
SARS-CoV-2 relevant PROSITE motifs.Chromatin is the tangled fibrous complex of DNA and protein within a eukaryotic nucleus.Protein-protein interactions (PPIs) are fundamental to the functioning of biological systems, playing a crucial role in essential processes such as cell signaling, . These include SMART and Pfam domains, as well as several protein . Lastly, MESSA is the most like GeneSilico as it provides information related to protein .SMART accession number: SM00165 Description: Present in Rad23, SNF1-like kinases.Prediction of domain homologues obviates misannotations of sequences that arise when comparing pairs of multidomain protein sequences with contrasting domain architectures.The domain architecture is defined as the linear order of all SMART domains in the protein sequence.Domain annotation has been updated with a detailed taxonomic breakdown and a prediction of the catalytic activity for 50 SMART domains is now available, based .Domain annotation has been updated with a detailed taxonomic breakdown and a prediction of the catalytic activity for 50 SMART domains is now available, based on the presence of essential amino acids. Nucleic Acids Res . Interpro abstract ( IPR017448 ): The scavenger receptor cysteine-rich (SRCR) domain is an ancient and . SMART (Simple Modular Architecture Research Tool) is a web resource ( https://smart. E -values have been the dominant statistic for protein sequence analysis for the past two decades: from identifying statistically significant local sequence alignments to evaluating matches to hidden Markov models describing protein domain families.de) for the identification and annotation of protein domains and the . This unit lists the range of tools available for domain prediction, and describes sequence and structural analysis tools that complement domain prediction methods.5 (gi 1938422), for example, is predicted by SMART to contain the following domains: S_TKc, S_TK_X, C1, CNH and .The Caenorhabditis elegans protein K08B12.SMART Domain Family Locale Probabilities. Nabil Ibtehaz, Yuki Kagaya & Daisuke Kihara.Protein domains are subunits that can fold and function independently., but just like PredictProtein, fails to deliver information about 3D-structure and function of the protein.
It includes protein domain and protein family models curated in house by CDD staff, as well as imported . A typical motif, such as a Zn-finger motif, is ten to twenty amino acids long. It is often associated with a distinct structural site performing a particular function.The January 2004 release of SMART contains 685 protein domains.SMART: a web-based tool for the study of genetically . New developments in SMART are centred on the integration of data from completed metazoan genomes.Protein domain databases remain important annotation and research tools. Interaction (with the environment) Molecules that sense cellular environmental change, such as osmolarity, light flux, acidity, ion concentration etc.These consensus sequence patterns are termed motifs and domains. GlobPlot: exploring protein sequences . The RCSB PDB also provides a variety of tools and resources. The last common ancestor of all organisms containing at least one protein with the domain architecture is defined as the point of its .
SMART: About SMART
Scavenger receptor Cys-rich. It is generated by mapping our genomic mode protein database .Along with the functional terms annotated per query, this new version of eggNOG-mapper provides PFAM (Mistry et al. This procedure also facilitates subsequent investigation of unannotated sequence regions thereby improving on the signal-to-noise ratio of the search .Domain-PFP allows protein function prediction using function-aware domain embedding representations. The SMART database ( 5) integrates manually curated hidden Markov models ( 6, 7) for . The Conserved Domain Database (CDD) is a freely available resource for the annotation of sequences with the locations of conserved protein domain footprints, as well as functional sites and motifs inferred from these footprints. Domain involved in innate immunity and lipid metabolism. The newly-found UBA in p62 is known to bind ubiquitin.
SMART: PH domain annotation
SMART version 9 contains manually curated models for more than 1300 protein domains, with a topical set of 68 new . The CDD's curated domain collections are often classified to a very . Also detailed are the basic domain prediction steps, . A protein can contain a single domain or multiple domains, each one typically associated with a specific function.Recognition and prediction of structural domains in proteins is an important part of structure and function prediction.Representation of a prediction of the amino acids in tertiary structures of homologues that overlay in three dimensions.Even though automated functional annotation of genes represents a fundamental step in most genomic and metagenomic workflows, it remains challenging at large scales. 2012 Jan;40(Database issue):D302-5. To derive the point of its invention, all proteins with the same domain architecture are mapped onto NCBIs taxonomy .Use Batch CD‐Search ( https://www.From Farm to Fork: Highlighting the Next Generation of Plant Proteins at the Smart Protein Closing Conference 2024. 2021) protein domain predictions.de) for the identification and annotation of protein domains .The backend of SMART is a PostgreSQL-based relational database management system, which stores the annotation of all SMART domains and the pre-calculated protein analyses for the entire Uniprot , Ensembl and STRING sequence databases.de) for the identification and annotation of protein domains and the analysis of protein domain architectures. Of 523 SMART (Schultz et al.Prediction of domain homologues obviates misannotations of sequences that arise when comparing pairs of multidomain protein sequences with contrasting domain . Interpro abstract ():UBA domains are a commonly occurring sequence motif of approximately 45 amino acid residues that are found in diverse proteins involved in the ubiquitin/proteasome pathway, DNA excision .SMART: Genomes available in Genomic SMARTsmart. 2021) and SMART (Letunic et al. Summary: Meta-DP, a domain prediction meta-server provides a simple interface to predict domains in a given protein sequence using a number of domain prediction methods.

Analysis of the secondary structure of MsIAA protein, the results found that, except for .Domains are considered to be the building blocks of protein structures.

The complete list of genomes in Genomic SMART .Accurate multiple alignments of 86 domains that occur in signaling proteins have been constructed and used to provide a Web-based tool (SMART: simple modular . Protein domain prediction Methods Mol Biol.

SMART (Simple Modular Architecture Research Tool) is an online resource ( http://smart.embl-heidelberg.
SMART: UBA domain annotation
SMART offers a wide-range of predictions like transmembrane helices, signal peptides, low complexity regions, homologous structures, domain architecture, etc. The underlying protein databases were synchronized with UniProt (2), Ensembl (3) and STRING (4), doubling the total number of annotated domains and other protein features .

The protein database in Normal SMART has significant redundancy, even though identical proteins are removed.
SMART: About SMART
Structure Prediction
There are 2836 ML domains in 2744 proteins in SMART's nrdb database. Description: The sea ucrhin egg peptide speract contains 4 repeats of SR domains that contain 6 conserved cysteines. PROSITE consists of documentation entries describing .SMART version 7 contains manually curated models for 1009 protein domains, 200 more than in the p . The Meta-DP is a convenient resource because through accessing a single site, users automatically obtain the results of the various domain prediction .toolsRecommandé pour vous en fonction de ce qui est populaire • Avis
SMART: Main page
Here we formally show that for “stratified” multiple hypothesis testing problems .The underlying protein databases were synchronized with UniProt (2), Ensembl (3) and STRING (4), doubling the total number of annotated domains and other protein features to more than 200 million. A motif is a short conserved sequence pattern associated with distinct functions of a protein or DNA. Recognition and prediction of structural domains in proteins is an important part of structure and function prediction.gov/Structure/bwrpsb/bwrpsb. We combine protein signatures from a number . Users can perform simple and advanced searches based on annotations relating to sequence, structure and function.

These methods are introduced in detail, .
PredictProtein
Perform SMART/Pfam domain prediction Minimum domain length (peak width) Maximum linker length (join distance) Plot settings Show dy/dx plot Show raw plot Invert background colour Output CASP format Title/ID Smoothing settings 1st derivative Savitzky-Golay frame 2nd derivative Savitzky-Golay frame Reference.1093/nar/gkr931. The problem is particularly challenging for . As deep learning algorithms drive the progress in protein structure prediction, a lot remains to be studied at this merging . SMART accession number: SM00233. The combination of domains determines the function of the protein, its subcellular localization and the interacti .If you notice something not working as expected, please contact us at help@predictprotein. There has been some progress in constructing useful models, especially for .SMART version 8 contains manually curated models for more than 1300 protein domains, with approximately 100 new models added since our last update article (1).Scientific Reports 10, Article number: 13374 ( 2020 ) Cite this article.SMART (Simple Modular Architecture Research Tool) is a web resource (http://smart.Motivation: Recognition models for protein-DNA interactions, which allow the prediction of specificity for a DNA-binding domain based only on its sequence or the alteration of specificity through rational design, have long been a goal of computational biology.Database of protein domains, families and functional sites. These molecules are visualized, .de/) for the identification and annotation of protein domains and the .